Particle Simulation
This project serves as my initial step toward scaling up simulations to analyze complex chemical systems. By refining accuracy and scalability, I aim to eventually explore molecular interactions, phase transitions, and reaction dynamics.


January 13 - 17, 2025
MD_SIM-AD-1-1
MD_SIM-AD-1-2

MD_SIM-AD-1-3

Successfully implemented randomized 3D particle initialization, pairwise force computation, and animated visualization of particle interactions. The model currently utilizes explicit Euler integration and a cutoff radius for computational efficiency. However, particles do not yet bounce off boundaries, as periodic boundary conditions have not been implemented. Future improvements include velocity Verlet integration for better energy conservation, implementing periodic boundary conditions or reflective walls, and optimizing force calculations using neighbor lists.
Continued with an extended version of a Lennard-Jones potential simulation, incorporating Manim for high-quality animation of molecular dynamics. Retained key components such as randomized 3D particle initialization, pairwise force computation, and Lennard-Jones interactions with a cutoff radius for efficiency. Still, particles do not experience periodic boundary conditions.
Further refined the Lennard-Jones potential simulation, enhancing Manim-based visualization by implementing trajectory tracking to capture particle motion over time. Maintained key features such as randomized 3D particle initialization, pairwise force computation, and Lennard-Jones interactions with a cutoff radius for efficiency. Reduced the number of particles to improve clarity in visualization. The code does apply periodic boundary conditions (PBCs) by wrapping particles inside the simulation box, but it does not correct forces across boundaries, which causes unrealistic jumps (as seen in the Manim visualization)
Progress Log:
> Challenge:
The animation initially used Matplotlib, but errors related to indexing and frame handling caused instability when rendering particle trajectories over multiple frames.
>> Solution:
After several debugging cycles, switched to Manim, which provided better frame-based rendering and avoided array indexing issues over time.
>Challenge:
The default rendering speed made the animation too fast, affecting interpretability.
>> Solution:
Increased `run_time` for each animation step and fine-tuned the frame rate to match expected motion speeds
Published on Github (Feb 11, 2025)
Feb 12, 2025
MD_SIM-AD-2-1
Further refined the Lennard-Jones potential simulation, improving boundary conditions, collision handling, and energy conservation while maintaining computational efficiency. This version builds upon MD_SIM-AD-1-3 by ensuring particles remain confined in the simulation box, reducing unrealistic velocity changes, and providing better visualization clarity.
Resources
"Understanding Molecular Simulation: from Algorithms to Applications"
Daan Frenkel & Berend Smit
"High Performance Python, Second Edition"
Micha Gorelick & Ian Ozsvald
"Desmond Users Guide"
D. E. Shaw Research
"Desmond 2.2 Users Guide"
Schrödinger Press


Reflective Boundary Conditions for Bouncing Particles
> Issue in MD_SIM-AD-1-3:
Particles escaped the simulation box.
>>Update:
Implemented reflective boundaries, where a particle’s velocity reverses upon hitting a boundary, keeping them inside the simulation box.


> Issue in MD_SIM-AD-1-3:
Particle collisions caused unrealistic velocity increases due to cumulative force effects.
>>Update:
Applied momentum conservation principles to ensure physically accurate velocity changes after collisions.
Improved Particle Collisions


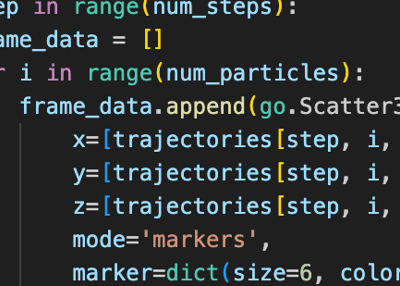
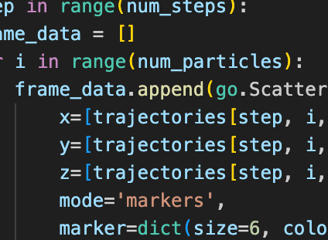
Velocity-Verlet Integration Optimization
> Issue in MD_SIM-AD-1-3:
Required improved numerical stability and energy conservation to model longer simulation.
>>Update:
Reduces drifting energy over long simulations and maintains a stable system.
Enhanced 3D Visualization
> Issue in MD_SIM-AD-1-3:
Limited rendering capacity and sub-optimal 3D visualization and render times via Manim
>>Update:
Implemented smooth trajectory lines to visualize particle motion over time, increased run_time per frame to improve interpretability, resolved indexing errors that caused instability in frame handling when initially trying to use matplotlib.